What is the NCGC Pharmaceutical Collection (NPC)?
The NCGC Pharmaceutical Collection (NPC) is a comprehensive,
publically-accessible collection of approved and
investigational drugs for high-throughput screening that
provides a
valuable resource for both validating new models of
disease and better understanding the molecular basis of
disease pathology and intervention. The NPC has already
generated several useful probes for studying a diverse
cross section of biology, including novel targets and
pathways. NCGC provides access to its set of approved
drugs and bioactives through the Therapeutics for Rare and Neglected Diseases (TRND) program and as part
of the compound collection for the
Tox21
initiative, a collaborative effort
for toxicity screening among several government agencies
including the US Environmental Protection Agency (EPA),
the National Toxicology Program (NTP), the US Food and
Drugs Administration (FDA), and the NCGC. Of
the nearly 2750 small molecular entities (MEs) that have been
approved for clinical use by US (FDA), EU (EMA),
Japanese (NHI), and Canadian (HC) authorities and that
are amenable to HTS screening, we currently possess 2,400
as part of our screening collection.
How do I get access to the NPC?
The NPC resource currently consists of (i) the physical collection suitable for high throughput screening (HTS) and (ii) the informatics browser and database. Putting together the physical collection has been surprisingly challenging in terms of the time and effort required in the informatics, compound management and synthetic chemistry related activities required for this endeavor. We provide access to the NPC screening library through collaboration. Please contact Noel Southall for additional information.The other half of the NPC resource is the NPC browser. This is a
self-contained software that is actively developed and maintained
by the informatics group to provide electronic access to the NPC content.
The latest version of the NPC browser for various platforms can be
downloaded below. Please let us know if your
platform is not listed. Note that a fairly modern hardware (preferably
with at least 2Gb of memory) is required to run the browser effectively.
How do I download the NPC browser?
| File | Platform | Size | MD5 checksum |
|---|---|---|---|
npc-browser_windows_1_2_1.exe |
Windows | 82 MB |
3335aeaf6326307eab1363cd22aa15da |
npc-browser_macos_1_2_1.dmg |
Mac OS X | 83 MB |
b128197c7a457183a8963fdb94087276 |
npc-browser_unix_1_2_1.sh |
Linux | 82 MB |
705e4df66fd1438595f1d88cdf5af796 |
Note that for the Mac OS X platform, it is assumed that Java 1.6 is already installed. If you're looking for 64-bit support, please try one of the downloads below.
npc-browser_windows-x64_1_2_1.exe |
Windows 64-bit | 82 MB |
9bbef3ab79ff098b7c1048c1ceeacc40 |
npc-browser_unix-x64_1_2_1.sh |
Linux 64-bit | 82 MB |
318d0c4619038d55a0c325f518bcfb0c |
Screenshots
 |
 |
 |
What's in the NPC browser?
The primary focus of the NPC browser was initially on approved
molecular entities. In the current version, however, we have expanded
it to include clinical candidates as well as other useful tool molecules.
We hope to eventually reach a steady state whereby new molecular entities
registered with the USAN and INN are automatically incorporated into the browser.
How reliable are the compound records in the NPC browser?
Although we strive to be as accurate as possible, due to the inherent
nature of how chemical structures are defined and represented electronically,
it is simply not possible to have an error-free database. Despite our best
efforts on curation, every structure is suspect until proven otherwise. This
sentiment certainly applies equally to any chemical database.
The NPC resource is—and will continue to be—an ongoing effort as
curation proceeds, new molecular entities are added as they are registered
or approved, and errors are found. This process will benefit enormously from
community feedback, and we encourage users to use the curation mechanism
within the browser to fix and/or report errors.
How can I build my own NPC physical collection?
One of the most challenging efforts in putting together the NPC physical
collection has been that of finding suitable and reliable chemical suppliers.
Furthermore, when drugs cannot be sourced, access to synthetic chemistry
is essential. Additionally, access to good compound management and
analytical chemistry resources is essential in quality control and proper
handling of the drugs collection. Where possible, we have included supplier
names and catalog identifiers in the browser to enable anyone to build their
own physical collection. Given the current collection has been sourced from
a combination of traditional chemical suppliers, specialty collections,
pharmacies and custom synthesis, any serious attempts at replicating the
NPC physical collection will likely to require significant amount of time
and resources.
Has the NPC screening library been characterized analytically?
The NCGC analytical chemistry group is currently generating QC LC/MS
data for all compounds in the NPC physical collection.
Where QC report is available,
a compound record in the NPC browser is marked with the icon
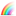 .
.
How do I cite the NPC resource?
If you make use of the NPC browser and/or database in your research, please use the following citation to acknowledge the NPC resource.R. Huang, N. Southall, Y. Wang, A. Yasgar, P. Shinn, A. Jadhav, D.-T. Nguyen, C. P. Austin, The NCGC Pharmaceutical Collection: A Comprehensive Resource of Clinically Approved Drugs Enabling Repurposing and Chemical Genomics. Sci. Transl. Med. 3, 80ps16 (2011).
Contact
Questions regarding collaboration or access to the NPC collection should be addressed to Ruili Huang and Noel Southall.Acknowledgements
The NPC resource has benefited immensely from community feedback since its initial release. We are particularly grateful to the following individuals who have generously donated their time and/or resources in helping us improve the NPC software and database:- Tudor Oprea and Oleg Ursu graciously donated the drug subset of the WOMBAT database. This dataset was instrumental in enabling us to validate a large number of curated structures (i.e., curation of curations so to speak).
- Manish Sud was an early adopter of the NPC resource. His thorough analysis helped us debugged a number of errors in the software as well as database.
- Matthew Hall provided valuable feedback on the handling of metal containing compounds (certainly beyond organometallics).
- Antony Williams' critical scrutinies of the compound content revealed numerous errors in the original version of the database. He also provided valuable feedback on other issues related to the software and data curation.
Change Log
| Ver. | Date | Changes |
|---|---|---|
| 1.2.0 | Feb 28, 2012 |
|
| 1.1.0 | June 28, 2011 |
|
| 1.0.23 | June 16, 2011 |
|
| 1.0.22 | June 6, 2011 |
|
| 1.0.21 | June 6, 2011 |
|
| 1.0.20 | May 8, 2011 | Minor bug fixes |
| 1.0.19 | May 3, 2011 |
|
| 1.0.18 | April 25, 2011 | Minor bug fixes |
| 1.0.17 | April 19, 2011 |
|
| 1.0.16 | April 4, 2011 | Enable double-clicking on any fragments and topologies to do implicit structure searching (i.e., performing substructure search without graph matching). |
| 1.0.15 | March 29, 2011 | |
| 1.0.14 | March 18, 2011 |
|
| 1.0.13 | February 25, 2011 |
|