Molecular framework forms the basis for which compounds are organized, indexed, and visualized within Tripod. There are two types of framework currently being generated when compounds are registered in Tripod: Fragment and topology. Here molecular fragments are derived based on exhaustive enumeration (up to some imposed limits) of all possible combinations of smallest set of smallest rings (SSSR). For a molecule that contains
k SSSR, the maximum number of possible fragments generated is in the order of 2
k - 1. In practice, however, the actual number of fragments generated for a typical small molecule is much lower due to symmetries and/or any number of additional constraints, e.g., synthetic accessibility/availability, reactivity filters, etc.
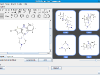
Consider, for example, the following molecule with its associated atom numbering.

The generated fragments are as follows:

Note that the so-called Murcko fragment (
2) is a byproduct of the enumeration. Even though there are only 5 unique fragments, we explicitly retain the atom mapping of the parent molecule as distinct fragments. This provides much flexibility for downstream analysis.
Molecular topologies are simply carbon skeletons of the molecular fragments. Below are the generated topologies for the given molecule.

Once generated, molecular fragments are extremely useful in many common tasks in cheminformatics. Our automated
R-group analysis tool is one example of such task. Another interesting use of fragments is for speeding up maximum common substructure (MCS) searches. That is, using fragments as initial seeds, we were able to significantly improve (an order of magnitude) on a branch-and-bound MCS algorithm. From clustering to 2D multi-graph alignment, we therefore see no limits to the utility of fragment-based approaches in cheminformatics. Coupled this sentiment with the recent progress in fragment-based screening, perhaps fragments are the future of drug discovery.
The tool to generate molecular fragments and topologies is available
here. Please feel free to let us know if you have any problems running it. As always, we appreciate any feedback.
 The generated fragments are as follows:
The generated fragments are as follows:
 Note that the so-called Murcko fragment (2) is a byproduct of the enumeration. Even though there are only 5 unique fragments, we explicitly retain the atom mapping of the parent molecule as distinct fragments. This provides much flexibility for downstream analysis.
Molecular topologies are simply carbon skeletons of the molecular fragments. Below are the generated topologies for the given molecule.
Note that the so-called Murcko fragment (2) is a byproduct of the enumeration. Even though there are only 5 unique fragments, we explicitly retain the atom mapping of the parent molecule as distinct fragments. This provides much flexibility for downstream analysis.
Molecular topologies are simply carbon skeletons of the molecular fragments. Below are the generated topologies for the given molecule.
 Once generated, molecular fragments are extremely useful in many common tasks in cheminformatics. Our automated R-group analysis tool is one example of such task. Another interesting use of fragments is for speeding up maximum common substructure (MCS) searches. That is, using fragments as initial seeds, we were able to significantly improve (an order of magnitude) on a branch-and-bound MCS algorithm. From clustering to 2D multi-graph alignment, we therefore see no limits to the utility of fragment-based approaches in cheminformatics. Coupled this sentiment with the recent progress in fragment-based screening, perhaps fragments are the future of drug discovery.
The tool to generate molecular fragments and topologies is available here. Please feel free to let us know if you have any problems running it. As always, we appreciate any feedback.
Once generated, molecular fragments are extremely useful in many common tasks in cheminformatics. Our automated R-group analysis tool is one example of such task. Another interesting use of fragments is for speeding up maximum common substructure (MCS) searches. That is, using fragments as initial seeds, we were able to significantly improve (an order of magnitude) on a branch-and-bound MCS algorithm. From clustering to 2D multi-graph alignment, we therefore see no limits to the utility of fragment-based approaches in cheminformatics. Coupled this sentiment with the recent progress in fragment-based screening, perhaps fragments are the future of drug discovery.
The tool to generate molecular fragments and topologies is available here. Please feel free to let us know if you have any problems running it. As always, we appreciate any feedback.
One thought on “Molecular framework”